This page contains succinct summaries of recently published research in cancer epigenetics. The information posted here is frequently updated, so please bookmark this page and visit us often to read the latest Cancer Epigenetics news at your leisure.
Latest on cancer epigenetics techniques

Cancer epigenetics: Streamlining 5-methylcytosine sequencing. Popular DNA methylation sequencing techniques utilize the principle of bisulfite conversion of DNA samples prior to analysis. A new parallelized DNA bisulfite conversion platform streamlines high throughput DNA sample handling and methylation analysis. The method relies on a droplet microfluidic design that decreases the number of processing steps and facilitates rapid bisulfite conversion. Reagent handling is reduced to a series of particle translocations with no complex fluidic control, and is coupled to a standard MethyLight DNA methylation assay (Tza-Huei Wang and coll., Johns Hopkins University, Baltimore, MD, USA).
Latest on miRNA-mediated cancer epigenetics
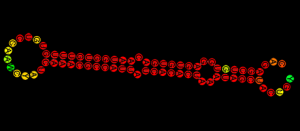
Cancer epigenetics: Self-assembled miRNA-gel scaffold is no miRage to cancer cells. RNA triple helices made up of a tumor suppressor miRNA and an antagomiR (oncomiR inhibitor) can be potent inhibitors of tumor growth in vitro. Dextran aldehyde can serve as an adhesive to tumor cells by interacting with natural tissue amines. Triplex conjugation of RNA helices to dendrimers yields stable triplex nanoparticles, which form an RNA-triple-helix gel scaffold following interaction with dextran aldehyde. Application of this gel to tumors leads to significant tumor shrinkage in a triple-negative breast cancer mouse model and doubled mouse median survival from 35 days to nearly 75 days. (João Conde, Natalie Artzi and coll., Harvard-MIT Division for Health Sciences and Technology, Cambridge, MA, USA).
Latest on histone-mediated cancer epigenetics
Cancer epigenetics: The autophagy-epigenetics strong link. mTOR (mechanistic target of rapamycin) serine/threonine kinase is a major regulator of autophagy. MTA2 (metastasis associated 1 family, member 2), a component of the nucleosome remodeling and histone deacetylase (NuRD) complex, recruits EZH2 to promoters of mTOR negative regulators (e.g. tuberous sclerosis 2 or TSC2), wherein it effects transcriptional repression. Down-regulation of TSC2 by EZH2 activates mTOR and inhibits autophagy. Expression of EZH2/MTA2 and that of TSC2 correlate negatively in colon carcinoma in patients, linking EZH2 to autophagy and tumor pathology (Wei-Guo Zhu, Ying Zhao and coll., Peking University Health Science Center, Beijing, China).
Latest on lncRNA-mediated cancer epigenetics
Cancer epigenetics: When TUG1 turns into a thug through excessive repression. Cyclin-dependent protein kinase inhibitors (CDKNs) keep the cell cycle in check to avoid uncontrolled cell growth. TUG1 lncRNA is overexpressed in gastric cancer wherein it binds PRC2. The pair binds to and trimethylates H3K27 at the p15, p16, p21, p27 and p57 CDKN gene loci, leading to their repression and to sustained cell growth. Knockdown of TUG1 reduces EZH2 binding to, and H3K27 trimethylation of all CDKN genes, with a concomitant derepression of all tested CDKNs, along with the inhibition of gastric tumor xenograft growth in mice. High TUG1 expression correlates to worse TNM stage and to metastasis in a cohort of 100 gastric cancer patients. Less than 40% of patients with high TUG1 expression versus more than 75% of patients with low TUG1 expression survive 36 months (Xu, Yin, De and coll., Nanjing Medical University, Jiangsu, China).